Background
The study and conservation of the natural world relies on detailed information about the distributions, abundances, and population trends of species over time. For many taxa, this information is challenging to obtain at relevant geographic scales. The goal of the eBird Status and Trends project is to use data from eBird, the global community science bird monitoring program administered by The Cornell Lab of Ornithology, to generate a reliable, standardized source of biodiversity information for the world’s bird populations. To translate the eBird observations into robust data products, we use machine learning to fill spatiotemporal gaps, using local land cover descriptions derived from remote sensing data, while controlling for biases inherent in species observations collected by community scientists. See Fink et al. (2019) for more information about the analysis used to generate these data.
This vignette gives an overview of the eBird Status Data Products, which estimate the full annual cycle distributions, relative abundances, and habitat associations for 2,980 species for the year 2023. For each species, distribution and abundance estimates are available for all 52 weeks of the year across a regular 3 km by 3 km square grid of cells covering the globe. Variation in detectability associated with the search effort is controlled by standardizing the estimates as the expected occurrence rate and count of the species on a 1 hour, 2 km checklist by an expert eBird observer at the optimal time of day and with optimal weather conditions for detecting the species.
Data access
Data access is granted through an Access Request Form at: https://ebird.org/st/request. Filling out this form generates a key to be used with this R package. Our terms of use have been designed to be quite permissive in many cases, particularly academic and research use. When requesting data access, please be sure to carefully read the terms of use and ensure that your intended use is not restricted.
After completing the Access Request Form, you will be provided an
eBird Status and Trends Data Products access key, which you will need
when downloading data. To store the key so this package can access it
when downloading data, use the function
set_ebirdst_access_key("XXXXX"), where "XXXXX"
is the access key provided to you.
There are a wide variety of data products available for download with
ebirdst via the function
ebirdst_download_status(). The first argument to this
function defines the species (as a common name, scientific name, or
species code) to download data for and the remaining arguments define
the specific data products to download. Throughout this vignettes, we’ll
use a simplified example dataset consisting of estimates for
Yellow-bellied Sapsucker in Michigan. This dataset is designed to be
small for faster download and is accessible without a key. By default
ebirst_download_status() only downloads the most commonly
used data products; however, since this vignette will cover all the
available data products, we’ll use download_all = TRUE.
Note that data for any species other that the example dataset requires a
key to access.
library(dplyr)
library(sf)
library(terra)
library(ebirdst)
# download a simplified example dataset for Yellow-bellied Sapsucker in Michigan
ebirdst_download_status(species = "yebsap-example", download_all = TRUE)By default, ebirdst_download_status() downloads data to
a centralized directory for on your computer. You can see what that
directory is with the function ebirdst_data_dir() and you
can change the default download directory by setting the environment
variable EBIRDST_DATA_DIR, for example by calling
usethis::edit_r_environ() and adding a line such as
EBIRDST_DATA_DIR=/custom/download/directory/.
IMPORTANT: eBird Status and Trends Data Products are designed
to be downloaded and accessed using the ebirdst R package.
Data downloaded using this R package have a specific file structure and
changing file names or locations will disrupt the ability of functions
in this package to access the data. If you prefer to access data for use
outside of R, consider downloading data via the eBird
Status and Trends website.
Species list
The data frame ebirdst_runs lists all species with eBird
Status Data Products available for download.
glimpse(ebirdst_runs)
#> Rows: 2,981
#> Columns: 30
#> $ species_code <chr> "yebsap-example", "abetow", "absfin1", …
#> $ scientific_name <chr> "Sphyrapicus varius", "Melozone aberti"…
#> $ common_name <chr> "Yellow-bellied Sapsucker", "Abert's To…
#> $ is_resident <lgl> FALSE, TRUE, TRUE, FALSE, TRUE, TRUE, F…
#> $ breeding_quality <chr> "3", NA, NA, "3", NA, NA, "1", NA, NA, …
#> $ breeding_start <date> 2023-05-17, NA, NA, 2023-05-31, NA, NA…
#> $ breeding_end <date> 2023-08-16, NA, NA, 2023-08-02, NA, NA…
#> $ nonbreeding_quality <chr> "3", NA, NA, "3", NA, NA, "1", NA, NA, …
#> $ nonbreeding_start <date> 2023-11-22, NA, NA, 2023-11-22, NA, NA…
#> $ nonbreeding_end <date> 2023-03-08, NA, NA, 2023-02-22, NA, NA…
#> $ postbreeding_migration_quality <chr> "3", NA, NA, "3", NA, NA, "0", NA, NA, …
#> $ postbreeding_migration_start <date> 2023-08-23, NA, NA, 2023-08-09, NA, NA…
#> $ postbreeding_migration_end <date> 2023-11-15, NA, NA, 2023-11-15, NA, NA…
#> $ prebreeding_migration_quality <chr> "3", NA, NA, "3", NA, NA, "0", NA, NA, …
#> $ prebreeding_migration_start <date> 2023-03-15, NA, NA, 2023-03-01, NA, NA…
#> $ prebreeding_migration_end <date> 2023-05-10, NA, NA, 2023-05-24, NA, NA…
#> $ resident_quality <chr> NA, "3", "3", NA, "3", "3", NA, "2", "3…
#> $ resident_start <date> NA, 2023-01-04, 2023-01-04, NA, 2023-0…
#> $ resident_end <date> NA, 2023-12-27, 2023-12-27, NA, 2023-1…
#> $ status_version_year <dbl> 2023, 2023, 2023, 2023, 2023, 2023, 202…
#> $ has_trends <lgl> TRUE, TRUE, FALSE, TRUE, TRUE, FALSE, F…
#> $ trends_season <chr> "breeding", "resident", NA, "breeding",…
#> $ trends_region <chr> "north_america", "north_america", NA, "…
#> $ trends_start_year <dbl> 2012, 2012, NA, 2012, 2011, NA, NA, NA,…
#> $ trends_end_year <dbl> 2022, 2022, NA, 2022, 2021, NA, NA, NA,…
#> $ trends_start_date <chr> "05-24", "01-25", NA, "05-24", "11-01",…
#> $ trends_end_date <chr> "08-16", "05-10", NA, "08-02", "05-03",…
#> $ rsquared <dbl> 0.8572896, 0.9231821, NA, 0.8570363, 0.…
#> $ beta0 <dbl> 0.227000849, -0.013923012, NA, 0.689424…
#> $ trends_version_year <dbl> 2022, 2022, NA, 2022, 2022, NA, NA, NA,…If you’re working in RStudio, you can use View() to
interactively explore this data frame.
All species go through a process of review by an expert on that
species prior to being released. The ebirdst_runs data
frame contains information from this review process. For migrants,
reviewers assess the model estimates for each of the four seasons:
breeding, non-breeding, pre-breeding migration, and post-breeding
migration. Resident (i.e., non-migratory) species are identified by
having TRUE in the is_resident column of
ebirdst_runs, and these species are assessed across the
whole year rather than seasonally. ebirdst_runs contains
two important pieces of information for each season: a
quality rating and seasonal dates.
The seasonal dates define the weeks that fall within each season. Breeding and non-breeding season dates are defined for each species as the weeks during those seasons when the species’ population does not move. For this reason, these seasons are also described as stationary periods. Migration periods are defined as the periods of movement between the stationary non-breeding and breeding seasons. Note that for many species these migratory periods include not only movement from breeding grounds to non-breeding grounds, but also post-breeding dispersal, molt migration, and other movements.
Reviewers also examine the model estimates for each season to assess the amount of extrapolation or omission present in the model, and assign an associated quality rating ranging from 0 (lowest quality) to 3 (highest quality). Extrapolation refers to cases where the model predicts occurrence where the species is known to be absent, while omission refers to the model failing to predict occurrence where a species is known to be present.
A rating of 0 implies this season failed review and model results should not be used at all for this period. Ratings of 1-3 correspond to a gradient of more to less extrapolation and/or omission, and we often use a traffic light analogy when referring to them:
- Red light (1): low quality, extensive extrapolation and/or omission and noise, but at least some regions have estimates that are accurate; can be used with caution in certain regions.
- Yellow light (2): medium quality, some extrapolation and/or omission; use with caution.
- Green light (3): high quality, very little or no extrapolation and/or omission; these seasons can be safely used.
Let’s look at the results of the review for our example dataset.
ebirdst_runs |>
filter(species_code == "yebsap-example") |>
glimpse()
#> Rows: 1
#> Columns: 30
#> $ species_code <chr> "yebsap-example"
#> $ scientific_name <chr> "Sphyrapicus varius"
#> $ common_name <chr> "Yellow-bellied Sapsucker"
#> $ is_resident <lgl> FALSE
#> $ breeding_quality <chr> "3"
#> $ breeding_start <date> 2023-05-17
#> $ breeding_end <date> 2023-08-16
#> $ nonbreeding_quality <chr> "3"
#> $ nonbreeding_start <date> 2023-11-22
#> $ nonbreeding_end <date> 2023-03-08
#> $ postbreeding_migration_quality <chr> "3"
#> $ postbreeding_migration_start <date> 2023-08-23
#> $ postbreeding_migration_end <date> 2023-11-15
#> $ prebreeding_migration_quality <chr> "3"
#> $ prebreeding_migration_start <date> 2023-03-15
#> $ prebreeding_migration_end <date> 2023-05-10
#> $ resident_quality <chr> NA
#> $ resident_start <date> NA
#> $ resident_end <date> NA
#> $ status_version_year <dbl> 2023
#> $ has_trends <lgl> TRUE
#> $ trends_season <chr> "breeding"
#> $ trends_region <chr> "north_america"
#> $ trends_start_year <dbl> 2012
#> $ trends_end_year <dbl> 2022
#> $ trends_start_date <chr> "05-24"
#> $ trends_end_date <chr> "08-16"
#> $ rsquared <dbl> 0.8572896
#> $ beta0 <dbl> 0.2270008
#> $ trends_version_year <dbl> 2022From this, we can see that Yellow-bellied Sapsucker was modeled as a migrant and all four seasons received a quality of 3, the highest rating. Note that there are a variety of trends-specific columns at the end of this data frame that we’ll ignore for now; these columns will be covered in the trends vignette
Data types
For each species, there are a variety of data products available, which can be categorized into the following broad types:
- Weekly raster estimates: weekly estimates of occurrence, count, relative abundance, and proportion of population on a regular grid in GeoTIFF format at three resolutions. These are the core products from which the other products are derived.
-
Seasonal raster estimates: seasonal estimates of
occurrence, count, relative abundance, and proportion of population on a
regular grid in GeoTIFF format at three resolutions. These are derived
from the corresponding weekly raster data by summarizing across the
weeks falling within each season based on the dates defined in the
ebirdst_runsdata frame. Only seasons that passed the expert review process are included. - Seasonal range boundaries: seasonal range boundary polygons in GeoPackage format.
- Regional summary statistics: a variety of summary statistics for countries and states/provinces (e.g. proportion of total population in the region) in CSV format.
- Predictive performance metrics (PPMs): a suite of spatial predictive performance metrics on a regular 27 km by 27 km grid in GeoTIFF format.
Each of these data products will be covered in more detail in the
following sections, including details on how to load the data into R.
All of the loading functions take a species (given as common name,
scientific name, or species code) as their first argument. If you have
used a non-default path argument to
ebirdst_download_status() then you will also need to
provide the same path argument to the loading
functions.
Weekly raster estimates
The core raster data products are the weekly estimates of occurrence, count, and relative abundance. These estimates are derived from an ensemble model producing 100 individual estimates of the expected value of each quantity. The raster data products give the median value across the ensemble for each quantity.
The weekly estimates are all stored in the widely used GeoTIFF raster
format, and we refer to them as “weekly cubes” (e.g. the “weekly
abundance cube”). All cubes have 52 weeks and cover the entire globe,
even for species with ranges only covering a small region. They come
with areas of predicted and assumed zeroes, such that any cells that are
NA represent areas where we didn’t produce model
estimates.
All estimates are the ensemble median expected value for a 2 km, 1 hour eBird Traveling Count by an expert eBird observer at the optimal time of day and for optimal weather conditions to observe the given species.
- Occurrence: the expected probability of encountering a species.
- Count: the expected count of a species, conditional on its occurrence at the given location.
- Relative abundance: the expected relative abundance of a species, computed as the product of the probability of occurrence and the count conditional on occurrence. In addition to the median relative abundance, upper and lower confidence intervals (CIs) are provided, defined at the 10th and 90th quantile of relative abundance, respectively.
- Proportion of population: the proportion of the total relative abundance within each cell. This is a derived product calculated by dividing each cell value in the relative abundance raster by the sum of all cell values
All predictions are made on a standard 3 km by 3 km global grid; however, for convenience lower resolution GeoTIFFs are also provided, which are typically much faster to work with. However, note that to keep file sizes small, the example dataset only contains lowest (27 km) resolution data. The three resolutions are:
- High resolution (3km): the native 3 km resolution data.
- Medium resolution (9km): the 3 km resolution data aggregated by a factor of 3 in each direction resulting in a resolution of 9 km.
- Low resolution (27km): the 3 km resolution data aggregated by a factor of 9 in each direction resulting in a resolution of 27 km.
The function load_raster() is used to load these data
into R and takes arguments for product and
resolution. The metric argument can be also be
used to access the relative abundance CIs. All raster products are
loaded into R as SpatRaster objects for use with the
terra R package. For example,
# weekly, 27km res, median relative abundance
abd_lr <- load_raster("yebsap-example", product = "abundance",
resolution = "27km")
# weekly, 27km res, median proportion of population
prop_pop_lr <- load_raster("yebsap-example", product = "proportion-population",
resolution = "27km")
# weekly, 27km res, abundance confidence intervals
abd_lower <- load_raster("yebsap-example", product = "abundance", metric = "lower",
resolution = "27km")
abd_upper <- load_raster("yebsap-example", product = "abundance", metric = "upper",
resolution = "27km")Each object has 52 layers, one for each week of the year, and layer names store the dates corresponding to the midpoints of each week.
as.Date(names(abd_lr))
#> [1] "2023-01-04" "2023-01-11" "2023-01-18" "2023-01-25" "2023-02-01"
#> [6] "2023-02-08" "2023-02-15" "2023-02-22" "2023-03-01" "2023-03-08"
#> [11] "2023-03-15" "2023-03-22" "2023-03-29" "2023-04-05" "2023-04-12"
#> [16] "2023-04-19" "2023-04-26" "2023-05-03" "2023-05-10" "2023-05-17"
#> [21] "2023-05-24" "2023-05-31" "2023-06-07" "2023-06-14" "2023-06-21"
#> [26] "2023-06-28" "2023-07-05" "2023-07-12" "2023-07-19" "2023-07-26"
#> [31] "2023-08-02" "2023-08-09" "2023-08-16" "2023-08-23" "2023-08-30"
#> [36] "2023-09-06" "2023-09-13" "2023-09-20" "2023-09-27" "2023-10-04"
#> [41] "2023-10-11" "2023-10-18" "2023-10-25" "2023-11-01" "2023-11-08"
#> [46] "2023-11-15" "2023-11-22" "2023-11-29" "2023-12-06" "2023-12-13"
#> [51] "2023-12-20" "2023-12-27"The GeoTIFFs use the Equal Earth coordinate reference system, an equal area projection suitable for global mapping and analysis.
Seasonal raster estimates
The seasonal raster estimates are provided for the same set of
products and at the same three resolutions as the weekly estimates.
They’re derived from the weekly data by taking the cell-wise mean or max
across the weeks within each season. The seasonal boundary dates are
defined through a process of expert review of each species, and are
available in the data frame ebirdst_runs. Each season is
also given a quality score from 0 (fail) to 3 (high quality), and
seasons with a score of 0 are not provided.
The function load_raster(period = "seasonal") is used to
load these data into R and takes arguments for product,
metric and resolution. The data are loaded
into R as SpatRaster objects for use with the
terra package. For example,
# seasonal, 27km res, mean relative abundance
abd_seasonal_mean <- load_raster("yebsap-example", product = "abundance",
period = "seasonal", metric = "mean",
resolution = "27km")
# season that each layer corresponds to
names(abd_seasonal_mean)
#> [1] "breeding" "nonbreeding" "prebreeding_migration"
#> [4] "postbreeding_migration"
# just the breeding season layer
abd_seasonal_mean[["breeding"]]
#> class : SpatRaster
#> size : 618, 1276, 1 (nrow, ncol, nlyr)
#> resolution : 27000, 27000 (x, y)
#> extent : -17226000, 17226000, -8343000, 8343000 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / Equal Earth Greenwich (EPSG:8857)
#> source : yebsap-example_abundance_seasonal_mean_27km_2023.tif
#> name : breeding
#> min value : 0.000000
#> max value : 1.021968
# seasonal, 27km res, max occurrence
occ_seasonal_max <- load_raster("yebsap-example", product = "occurrence",
period = "seasonal", metric = "max",
resolution = "27km")Finally, as a convenience, the data products include year-round
rasters summarizing the mean or max across all weeks that fall within a
season that passed the expert review process. These can be accessed
similarly to the seasonal products, but with
period = "full-year" instead. For example, these layers can
be used in conservation planning to assess the most important sites
across the full range and full annual cycle of a species.
# full year, 27km res, maximum relative abundance
abd_fy_max <- load_raster("yebsap-example", product = "abundance",
period = "full-year", metric = "max",
resolution = "27km")Range boundaries
Seasonal range polygons are defined as the boundaries of non-zero
seasonal relative abundance estimates, which are then (optionally)
smoothed to produce more aesthetically pleasing polygons using the
smoothr package. They are provided in the widely used
GeoPackage format and can be loaded into R with
load_ranges(), which returns a set of spatial features for
use with the sf R package. By default the smoothed ranges
are returned, but using smoothed = FALSE will return the
raw, unsmoothed range polygons. Note that only low and medium resolution
ranges are provided. These range polygons can be loaded with
load_ranges():
# seasonal, 27km res, smoothed ranges
ranges <- load_ranges("yebsap-example", resolution = "27km")
ranges
#> Simple feature collection with 4 features and 8 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -90.41254 ymin: 41.69681 xmax: -82.4146 ymax: 48.19076
#> Geodetic CRS: WGS 84
#> # A tibble: 4 × 9
#> species_code scientific_name common_name prediction_year type season
#> <chr> <chr> <chr> <int> <chr> <chr>
#> 1 yebsap Sphyrapicus varius Yellow-bellied S… 2023 range breed…
#> 2 yebsap Sphyrapicus varius Yellow-bellied S… 2023 range nonbr…
#> 3 yebsap Sphyrapicus varius Yellow-bellied S… 2023 range postb…
#> 4 yebsap Sphyrapicus varius Yellow-bellied S… 2023 range prebr…
#> # ℹ 3 more variables: start_date <date>, end_date <date>,
#> # geom <MULTIPOLYGON [°]>
# subset to just the breeding season range using dplyr
range_breeding <- filter(ranges, season == "breeding")Regional summary statistics
Regional summaries of the seasonal raster estimates are also provided
for a standard set of regions (countries and states/provinces). These
summary statistics can be loaded with
load_regional_stats():
regional <- load_regional_stats("yebsap-example")
glimpse(regional)
#> Rows: 8
#> Columns: 15
#> $ species_code <chr> "yebsap-example", "yebsap-example", "yebsap-exa…
#> $ region_type <chr> "country", "country", "country", "country", "st…
#> $ region_code <chr> "USA", "USA", "USA", "USA", "USA-MI", "USA-MI",…
#> $ region_name <chr> "United States", "United States", "United State…
#> $ continent_code <chr> "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA"
#> $ continent_name <chr> "North America", "North America", "North Americ…
#> $ season <chr> "breeding", "nonbreeding", "postbreeding_migrat…
#> $ abundance_mean <dbl> 0.114652605, 0.123534772, 0.073477334, 0.084178…
#> $ total_pop_percent <dbl> 0.3034155366, 0.9497140025, 0.7885750146, 0.435…
#> $ continent_pop_percent <dbl> 0.3034155366, 0.9497140025, 0.7885750146, 0.435…
#> $ range_occupied_percent <dbl> 0.25990556, 0.40518814, 0.54434548, 0.49810429,…
#> $ range_total_percent <dbl> 0.231714761, 0.801986186, 0.720437099, 0.573623…
#> $ range_days_occupation <int> 98, 112, 91, 63, 98, 112, 91, 49
#> $ max_week <chr> "2023-08-16", "2023-11-22", "2023-10-18", "2023…
#> $ max_week_percent_pop <dbl> 0.4100934563, 0.9638256049, 0.9979910064, 0.985…The six summary statistics are defined as:
-
abundance_mean: mean relative abundance in the region. -
total_pop_percent: the proportion of the seasonal modeled population within the region. -
continent_pop_percent: the proportion of the seasonal modeled population for the continent within the region. Thecontinent_namecolumn identifies the continent that the region falls within. Note that Yellow-bellied Sapsucker only occurs in North American so the total and continental proportions are identical. -
range_percent_occupied: the proportion of the region occupied by the species during the given season. -
range_total_percent: the proportion of the species seasonal range falling within the region. -
range_days_occupation: number of days of the season that the region was occupied by this species.
Predictive performance metrics (PPMs)
A subset of 10% of the eBird observations is excluded from model training to be used as a test set. For each submodel making up the eBird Status model ensemble, model predictions are compared to actual occurrence and count for a spatiotemporally subsampled version of the test dataset to generate a suite of predictive performance metrics (PPMs) at the base model level. These PPMs are then summarized across the ensemble to a 27 km resolution raster grid, where the cell values are the average across all models in the ensemble contributing to that cell. For migrants, PPMs are provided at weekly temporal resolution, in the form of a stack of 52 rasters for each metric, while for residents, year round PPMs are provided in the form of a single raster for each metric.
In total, nineteen predictive performance metrics are provided in four categories. Binary PPMs compare the predicted presence/absence to observed detection/non-detection for all test checklists.
-
binary_f1: F1-score. -
binary_mcc: Matthews Correlation Coefficient (MCC). -
binary_prevalence: the observed detection probability after spatiotemporal subsampling.
Occurrence PPMs compare the predicted encounter rate with the observed detection/non-detection for the subset of test checklists within the predicted range boundary.
-
occ_bernoulli_dev: the proportion of the Bernoulli deviance explained. -
occ_bin_spearman: test observations are binned by predicted encounter rate with bin widths of 0.05, then the mean observed prevalence and predicted encounter rate are calculated within bins. This metric is the Spearman’s rank correlation coefficient comparing the observed and predicted binned mean values. -
occ_brier: Brier score, i.e. the mean squared difference between predicted encounter rate and observed detection/non-detection. -
occ_pr_auc: area on the precision-recall curve (PR-AUC) -
occ_pr_auc_gt_prev: the proportion of the ensemble for which the PR-AUC is greater than observed prevalence, which indicates that the model is performing better than random guessing. -
occ_pr_auc_normalized: the PR-AUC normalized to account for class imbalance so that a value of 0 represents performance equal to random guessing and a value of 1 represents perfect classification.
Count PPMs compare the predicted count with the observed count for the subset of test checklists within the predicted range boundary and where the species was detected.
-
count_log_pearson: the Pearson correlation coefficient comparing the logarithm of the predicted count with the logarithm of the observed count. -
count_mae: mean absolute error (MAE). -
count_poisson_dev: the proportion of the Poisson deviance explained. -
count_rmse: route mean squared error (RMSE). -
count_spearman: Spearman’s rank correlation coefficient.
Abundance PPMs compare the predicted relative abundance with the observed count for the full set of tests checklists.
-
abd_log_pearson: Pearson correlation coefficient comparing the logarithm of the predicted relative abundance with the logarithm of the observed count. -
abd_mae: the mean absolute error (MAE). -
abd_poisson_dev: the proportion of the Poisson deviance explained. -
abd_rmse: root mean squared error (RMSE). -
abd_spearman: Spearman’s rank correlation coefficient.
The spatial PPMs can be loaded using load_ppms(). For
example, to load the normalized PR-AUC for the example dataset use.
pr_auc <- load_ppm("yebsap-example", ppm = "occ_pr_auc_normalized")
print(pr_auc)
#> class : SpatRaster
#> size : 618, 1276, 52 (nrow, ncol, nlyr)
#> resolution : 27000, 27000 (x, y)
#> extent : -17226000, 17226000, -8343000, 8343000 (xmin, xmax, ymin, ymax)
#> coord. ref. : WGS 84 / Equal Earth Greenwich (EPSG:8857)
#> source : yebsap-example_ppm_occ-pr-auc-normalized_mean_27km_2023.tif
#> names : 01-04, 01-11, 01-18, 01-25, 02-01, 02-08, ...
#> min values : 0.1113430, 0.09812936, 0.0189133, 0.0189133, 0.0189133, 0.01403384, ...
#> max values : 0.7343874, 0.73438740, 0.7969337, 0.7969337, 1.0000000, 1.00000000, ...Since Yellow-bellied Sapsucker is a migrant, there are 52 layers, one
for each week of the year, giving the mean PR-AUC for each 27 km by 27
km grid cell. We can produce a simple plot of PR-AUC for a week in the
middle of the year. Note that trim() is used to trim the
global raster down to just show the area with data (the state of
Michigan in this example dataset).
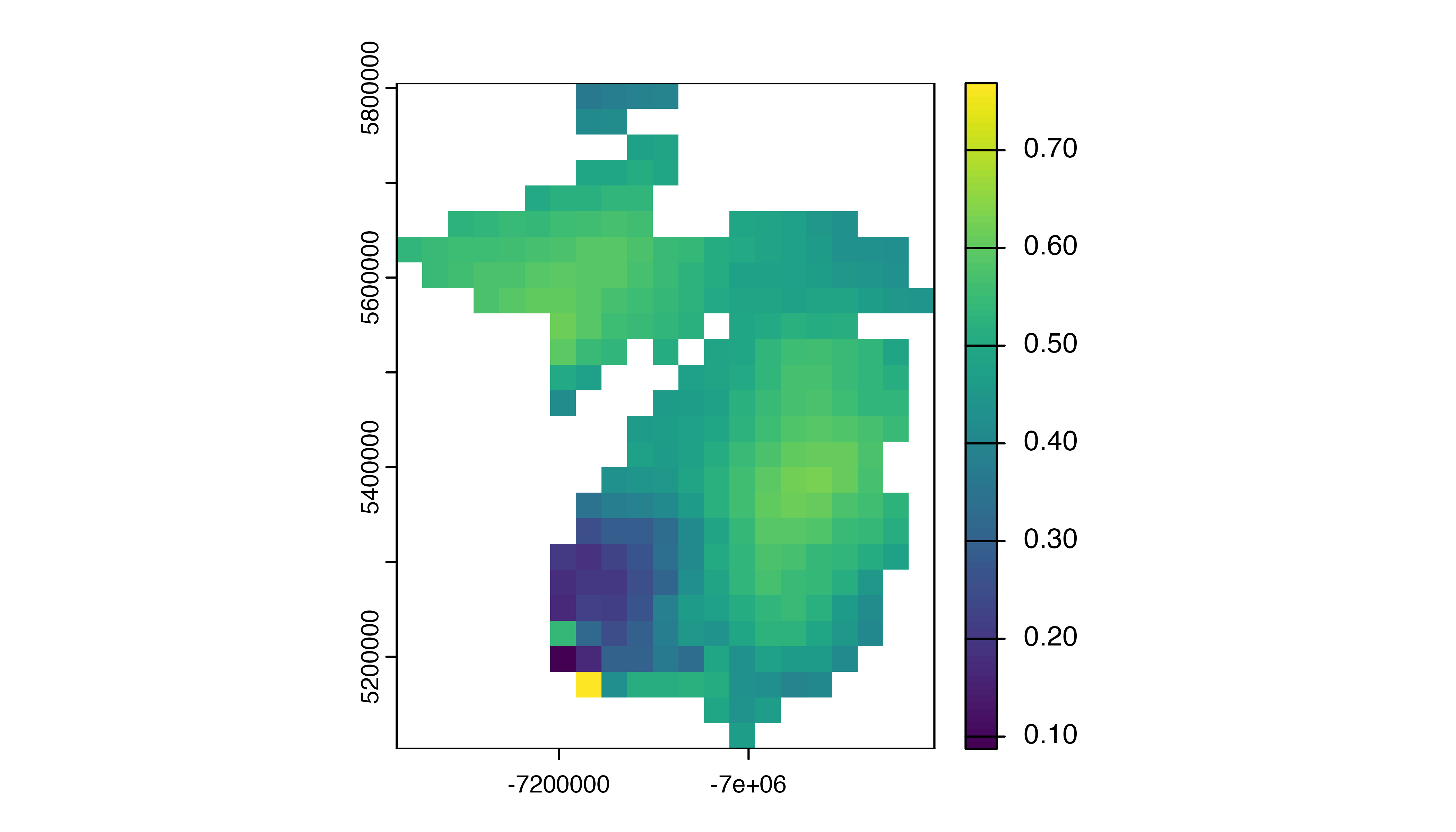
See the applications vignette for a more detailed example of how to use the PPMs.
Data coverage
In addition to the species-specific data products discussed above,
ebirdst provides access to two species agnostic data
products from out data coverage workflow. The data products in GeoTIFF
format provide weekly estimates on a regular 3 km by 3km grid of
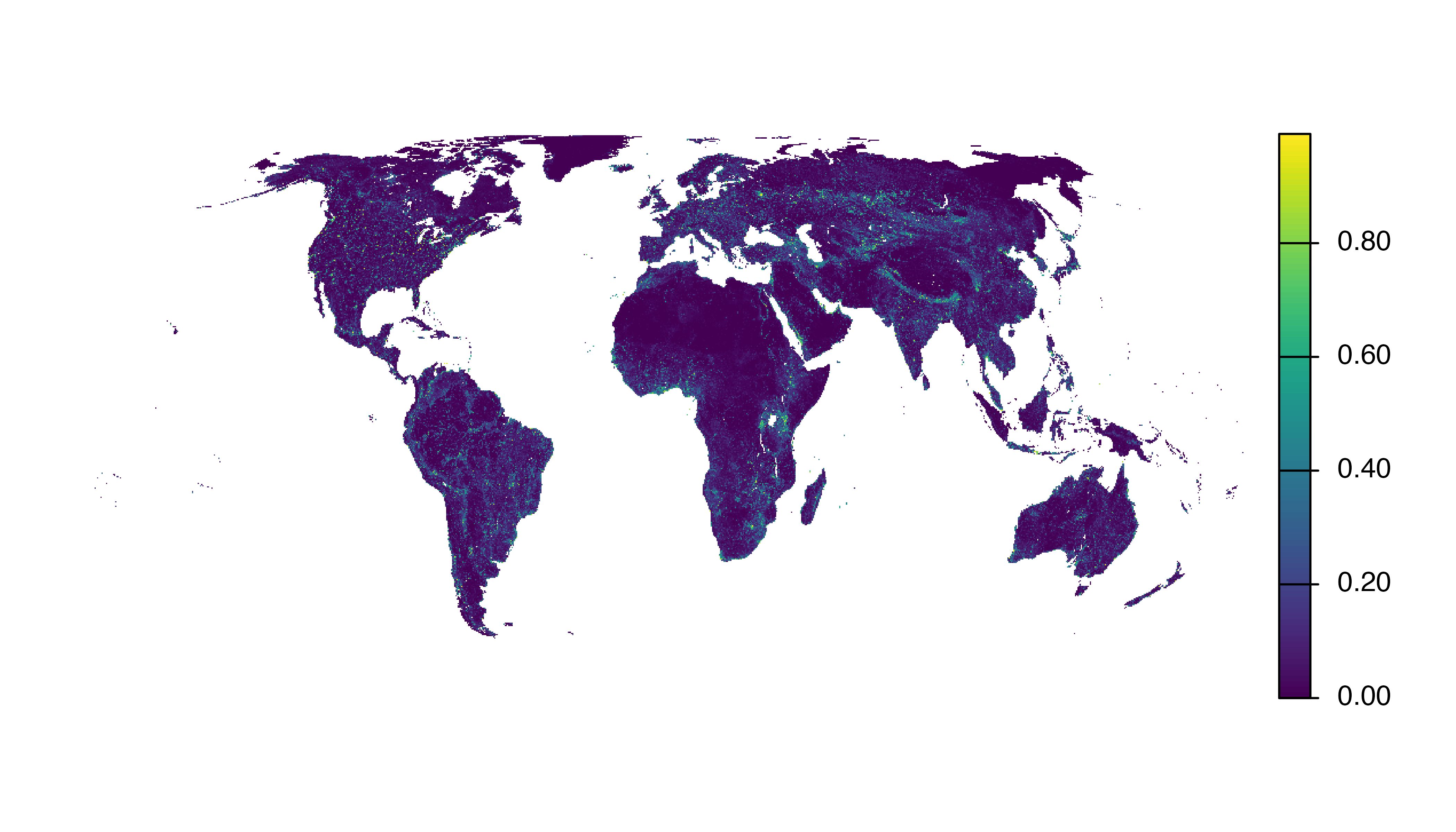
- Site selection probability: the modeled probability (0-1) that a grid cell of a certain habitat configuration received an eBird checklist within that region and season.
- Spatial coverage: the fraction (0-1) of grid cells within that region and season that checklists for the given week.
These data products identify areas where the coverage of eBird data is relatively high or low, which can be used to prioritize areas for increased data collection. To download the data coverage data products use:
Then to load the data, for example, site selection probability for the week of May 10, and make a map, use:
site_sel <- load_data_coverage("selection-probability", week = "05-10")
plot(site_sel, axes = FALSE)