library(dplyr)
library(ggplot2)
library(sf)
library(terra)
library(ebirdst)2 eBird Trends Data Products
The eBird Trends Data Products provide estimates of trends in relative abundance based on eBird data. Trend estimates are made on a 27 km by 27 km grid for a single season per species (breeding, non-breeding, or resident). For further details on the methodology used to estimate these trends consult the associated paper:
Fink, D., Johnston, A., Strimas-Mackey, M., Auer, T., Hochachka, W. M., Ligocki, S., Oldham Jaromczyk, L., Robinson, O., Wood, C., Kelling, S., & Rodewald, A. D. (2023). A Double machine learning trend model for citizen science data. Methods in Ecology and Evolution, 00, 1–14. https://doi.org/10.1111/2041-210X.14186
This lesson will introduce you to the eBird Trends Data Products. We will demonstrate how to download trends data, load the data into R, and use it to estimate regional trends and multi-species trends. Let’s start by loading the packages we’ll use throughout this lesson
2.1 eBird Trends species
The 2022 release of the eBird Trends Data Products includes data for 853 species. The data frame ebirdst_runs that we examined in the previous lesson indicates which species have trends estimates with the has_trends column. We can filter the data frame and only select those columns relevant to trends.
trends_runs <- ebirdst_runs %>%
filter(has_trends) %>%
select(species_code, common_name,
trends_season, trends_region,
trends_start_year, trends_end_year,
trends_start_date, trends_end_date,
rsquared, beta0)
glimpse(trends_runs)
#> Rows: 853
#> Columns: 10
#> $ species_code <chr> "abetow", "acafly", "acowoo", "aldfly", "alepar2", "…
#> $ common_name <chr> "Abert's Towhee", "Acadian Flycatcher", "Acorn Woodp…
#> $ trends_season <chr> "resident", "breeding", "resident", "breeding", "res…
#> $ trends_region <chr> "north_america", "north_america", "north_america", "…
#> $ trends_start_year <dbl> 2012, 2012, 2011, 2012, 2015, 2012, 2011, 2011, 2012…
#> $ trends_end_year <dbl> 2022, 2022, 2021, 2022, 2022, 2022, 2021, 2021, 2022…
#> $ trends_start_date <chr> "01-25", "05-24", "11-01", "06-21", "07-26", "03-22"…
#> $ trends_end_date <chr> "05-10", "08-02", "05-03", "07-12", "12-06", "06-07"…
#> $ rsquared <dbl> 0.923, 0.857, 0.881, 0.787, 0.820, 0.909, 0.852, 0.7…
#> $ beta0 <dbl> -0.01392, 0.68942, -0.09267, -0.16241, 0.31904, -0.1…Information is provided on the trends model for each species, including two predictive performance metrics (rsquared and beta0) that are based on a comparison of actual and estimated trends for a suite of simulations (see Fink et al. 2023 for further details). The columns in the trends_runs data frame are as follows:
species_code: the alphanumeric eBird species code uniquely identifying the species.common_name: the English common name of the species.trends_season: season that the trend was estimated for: breeding, nonbreeding, or resident.trends_region: the geographic region that the trend model was run for. Note that broadly distributed species (e.g. Barn Swallow) will only have trend estimates for a regional subset of their full range.trends_start_year/trends_end_year: the start and end years of the trend time period.trends_start_date/trends_end_date: the start and end dates (MM-DDformat) of the season for which the trend was estimated.rsquared: R-squared value comparing the actual and estimated trends from the simulations.beta0: the intercept of a linear model fitting actual vs. estimated trends (actual ~ estimated) for the simulations. Positive values ofbeta0indicate that the models are systematically underestimating the simulated trend for this species.
Note that some season dates span two calendar years, for example Canvasback has 2011-2021 trends estimates for a non-breeding season defined as December 20 to January 25. In this case, the first season will be December 20, 2011 to Janurary 25, 2012.
trends_runs %>%
filter(common_name == "Canvasback") %>%
select(trends_start_year, trends_end_year,
trends_start_date, trends_end_date)
#> # A tibble: 1 × 4
#> trends_start_year trends_end_year trends_start_date trends_end_date
#> <dbl> <dbl> <chr> <chr>
#> 1 2011 2021 12-20 01-252.2 Downloading data
Trends data access is granted through the same process as for the eBird Status Data Products. If you haven’t already requested an API key, consult the relevant section in the previous lesson.
Trends data can be downloaded for one or more species using ebirdst_download_trends(), where the first argument is a vector of common names, scientific names, or species codes. As with the Status Data Products, trends data will be downloaded to a centralized directory which can be viewed with ebirdst_data_dir(). For example, let’s download the breeding season trends data for Sage Thrasher.
ebirdst_download_trends("Sage Thrasher")2.3 Loading data into R
Once the data are downloaded, the trends data for a set of species, can be loaded into R using the function load_trends(). For example, we can load the Sage Thrasher trends estimates we just downloaded with:
trends_sagthr <- load_trends("Sage Thrasher")Each row corresponds to the trend estimate for a 27 km by 27 km grid cell, identified by the srd_id column and with cell center given by the longitude and latitude coordinates. Columns beginning with abd_ppy provide estimates of the percent per year trend in relative abundance and 80% confidence intervals, while those beginning with abd_trend provide estimates of the cumulative trend in relative abundance and 80% confidence intervals over the time period. The abd column gives the relative abundance estimate for the middle of the trend time period (e.g. 2014 for a 2007-2021 trend). The start_year/end_year and start_date/end_date columns provide redundant information to that available in ebirdst_runs. Specifically for Sage Thrasher we have:
trends_runs %>%
filter(common_name == "Sage Thrasher") %>%
select(trends_start_year, trends_end_year,
trends_start_date, trends_end_date)
#> # A tibble: 1 × 4
#> trends_start_year trends_end_year trends_start_date trends_end_date
#> <dbl> <dbl> <chr> <chr>
#> 1 2012 2022 05-17 07-12This tells us that the trend estimates are for the breeding season (May 17 to July 12) for the period 2012-2022.
2.4 Conversion to spatial formats
The eBird trends data are stored in a tabular format, where each row gives the trend estimate for a single cell on a 27 km x 27 km equal area grid. For each grid cell, the coordinates (longitude and latitude) are provided for the center of the grid cell. For many applications, an explicitly spatial format is more useful and these coordinates can be use to convert from the tabular format to either a vector or raster format.
2.4.1 Vector (points)
The tabular trends data can be converted into point vector features for use with the sf package using the sf function st_as_sf().
trends_sf <- st_as_sf(trends_sagthr,
coords = c("longitude", "latitude"),
crs = 4326)
print(trends_sf)
#> Simple feature collection with 2462 features and 15 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -122 ymin: 33.5 xmax: -103 ymax: 49.4
#> Geodetic CRS: WGS 84
#> # A tibble: 2,462 × 16
#> species_code season start_year end_year start_date end_date srd_id abd
#> * <chr> <chr> <int> <int> <chr> <chr> <int> <dbl>
#> 1 sagthr breeding 2012 2022 05-17 07-12 254264 0.000527
#> 2 sagthr breeding 2012 2022 05-17 07-12 255764 0.0147
#> 3 sagthr breeding 2012 2022 05-17 07-12 255765 0.000214
#> 4 sagthr breeding 2012 2022 05-17 07-12 257264 0.00174
#> 5 sagthr breeding 2012 2022 05-17 07-12 257265 0.0132
#> 6 sagthr breeding 2012 2022 05-17 07-12 257266 0.00118
#> # ℹ 2,456 more rows
#> # ℹ 8 more variables: abd_ppy <dbl>, abd_ppy_lower <dbl>, abd_ppy_upper <dbl>,
#> # abd_ppy_nonzero <lgl>, abd_trend <dbl>, abd_trend_lower <dbl>,
#> # abd_trend_upper <dbl>, geometry <POINT [°]>2.4.2 Raster
The tabular trend estimates can most easily be converted to raster format for use with the terra package using the function rasterize_trends(). Any of the columns in the trends data frame can be selected using the layers argument and converted into layers in the resulting raster object.
# rasterize the percent per year trend with confidence limits (default)
ppy_raster <- rasterize_trends(trends_sagthr)
print(ppy_raster)
#> class : SpatRaster
#> dimensions : 67, 100, 3 (nrow, ncol, nlyr)
#> resolution : 26665, 26665 (x, y)
#> extent : -10602273, -7935747, 3714548, 5501122 (xmin, xmax, ymin, ymax)
#> coord. ref. : +proj=sinu +lon_0=0 +x_0=0 +y_0=0 +R=6371007.181 +units=m +no_defs
#> source(s) : memory
#> names : abd_ppy, abd_ppy_lower, abd_ppy_upper
#> min values : -14.6, -17.5, -11.5
#> max values : 13.6, 11.7, 15.8
# rasterize the cumulative trend estimate
trends_raster <- rasterize_trends(trends_sagthr, layers = "abd_trend")
print(trends_raster)
#> class : SpatRaster
#> dimensions : 67, 100, 1 (nrow, ncol, nlyr)
#> resolution : 26665, 26665 (x, y)
#> extent : -10602273, -7935747, 3714548, 5501122 (xmin, xmax, ymin, ymax)
#> coord. ref. : +proj=sinu +lon_0=0 +x_0=0 +y_0=0 +R=6371007.181 +units=m +no_defs
#> source(s) : memory
#> name : abd_trend
#> min value : -79.4
#> max value : 258.9A simple map of these data can be produced from the raster data. For example, we’ll make a map of percent per year change in relative abundance for Sage Thrasher.
# define breaks and palettes similar to those on status and trends website
breaks <- seq(-4, 4)
breaks[1] <- -Inf
breaks[length(breaks)] <- Inf
pal <- ebirdst_palettes(length(breaks) - 1, type = "trends")
# make a simple map
plot(ppy_raster[["abd_ppy"]],
col = pal, breaks = breaks,
main = "Trend in relative abundance 2012-2022 [% change per year]",
cex.main = 0.75,
axes = FALSE)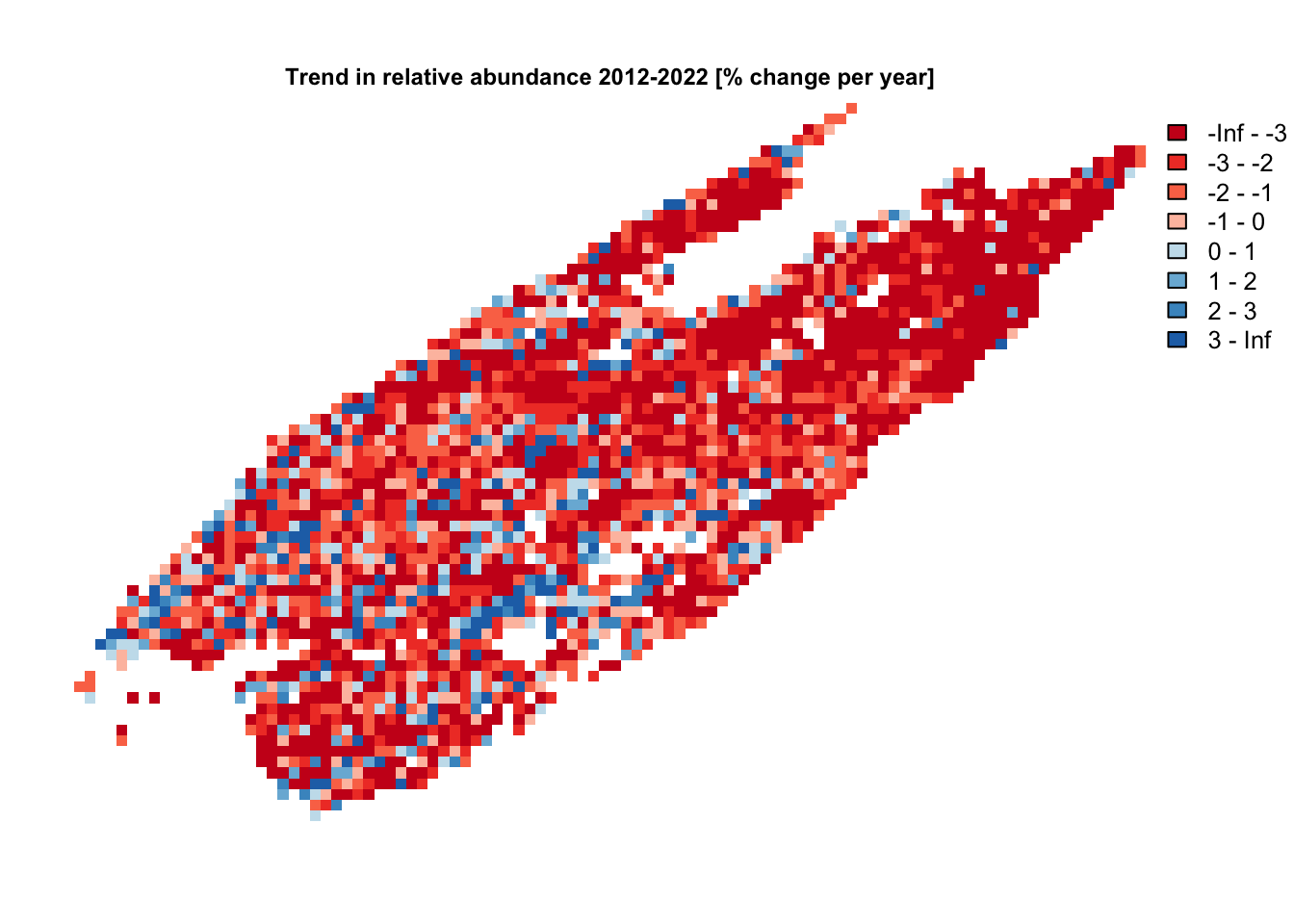
The above map shows the percent per year trend, which is different from the trends maps on the Status and Trends website that show cumulative trend. These are two different representation of the same data and we can convert between the two using the exponential growth formula. For example, a 1% annual decline equates to a -9.6% cumulative decline from 2012-2022.
trend_ppy <- -1
(trend_cumulative <- 100 * ((1 + trend_ppy / 100)^(2022 - 2012) - 1))
#> [1] -9.562.5 Uncertainty
Recall that the trends data frame contained estimates of uncertainty in the relative abundance trend given as 80% confidence intervals. How were these confidence intervals calculated? The model used to estimate trends produces an ensemble of 100 estimates for each grid cell, each based on a random subsample of eBird data. This ensemble of estimates is used to quantify uncertainty in the trends estimates. The estimated trend is the median across the ensemble, and the 80% confidence intervals are the lower 10th and upper 90th percentiles across the ensemble. Those wishing to access estimates from the individual folds making up the ensemble can use fold_estimates = TRUE when loading data. These fold-level estimates can be used to quantify uncertainty, for example, when calculating the trend for a given region. For example, let’s load the fold-level estimates for Sage Thrasher:
trends_sagthr_folds <- load_trends("sagthr", fold_estimates = TRUE)
print(trends_sagthr_folds)
#> # A tibble: 246,200 × 8
#> species_code season fold srd_id latitude longitude abd abd_ppy
#> <chr> <chr> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 sagthr breeding 1 254264 49.4 -120. 0.000527 -3.11
#> 2 sagthr breeding 1 255764 49.1 -120. 0.0147 -2.97
#> 3 sagthr breeding 1 255765 49.1 -119. 0.000214 -2.25
#> 4 sagthr breeding 1 257264 48.9 -120. 0.00174 -4.53
#> 5 sagthr breeding 1 257265 48.9 -120. 0.0132 -3.86
#> 6 sagthr breeding 1 257266 48.9 -119. 0.00118 -4.04
#> # ℹ 246,194 more rowsThis data frame is much more concise, only giving estimates of the mid-point relative abundance and percent per year trend in relative abundance for each of 100 folds for each grid cell.
2.6 Applications
2.6.1 Regional trends
eBird trends estimates are made on a 27 km by 27 km grid, which allows summarization over broader regions such as states or provinces. Since the relative abundance of a species varies throughout its range, we need to weight the mean trend calculation by relative abundance (abd in the trends data frame). To quantify uncertainty in the regional trend, we can use the fold-level data to produce 100 distinct estimates of the regional trend, then calculate the median and 80% confidence intervals. As an example, let’s calculate the state-level mean percent per year trends in relative abundance for Sage Thrasher.
# boundaries of states in the united states
states <- paste0("https://github.com/ebird/ebirdst-workshop_tws-2023/",
"raw/main/data/boundaries.gpkg") %>%
read_sf(layer = "states")
# convert fold-level trends estimates to sf format
trends_sagthr_sf <- st_as_sf(trends_sagthr_folds,
coords = c("longitude", "latitude"),
crs = 4326)
# attach state to the fold-level trends data
trends_sagthr_sf <- st_join(trends_sagthr_sf, states, left = FALSE)
# abundance-weighted average trend by region and fold
trends_states_folds <- trends_sagthr_sf %>%
st_drop_geometry() %>%
group_by(state, fold) %>%
summarize(abd_ppy = sum(abd * abd_ppy) / sum(abd),
.groups = "drop")
# summarize across folds for each state
trends_states <- trends_states_folds %>%
group_by(state) %>%
summarise(abd_ppy_median = median(abd_ppy, na.rm = TRUE),
abd_ppy_lower = quantile(abd_ppy, 0.10, na.rm = TRUE),
abd_ppy_upper = quantile(abd_ppy, 0.90, na.rm = TRUE),
.groups = "drop") %>%
arrange(abd_ppy_median)
head(trends_states)
#> # A tibble: 6 × 4
#> state abd_ppy_median abd_ppy_lower abd_ppy_upper
#> <chr> <dbl> <dbl> <dbl>
#> 1 US-SD -5.42 -8.36 -4.33
#> 2 US-MT -3.90 -5.33 -3.11
#> 3 US-WA -3.70 -4.48 -2.83
#> 4 US-ID -3.51 -4.19 -2.55
#> 5 US-AZ -3.44 -3.94 -2.25
#> 6 US-WY -3.42 -3.83 -2.51We can join these state-level trends back to the state boundaries and make a map with ggplot2.
trends_states_sf <- left_join(states, trends_states)
ggplot(trends_states_sf) +
geom_sf(aes(fill = abd_ppy_median)) +
scale_fill_distiller(palette = "Reds",
limits = c(NA, 0),
na.value = "grey80") +
guides(fill = guide_colorbar(title.position = "top", barwidth = 15)) +
labs(title = "Sage Thrasher state-level breeding trends 2012-2022",
fill = "Relative abundance trend [% change / year]") +
theme_bw() +
theme(legend.position = "bottom")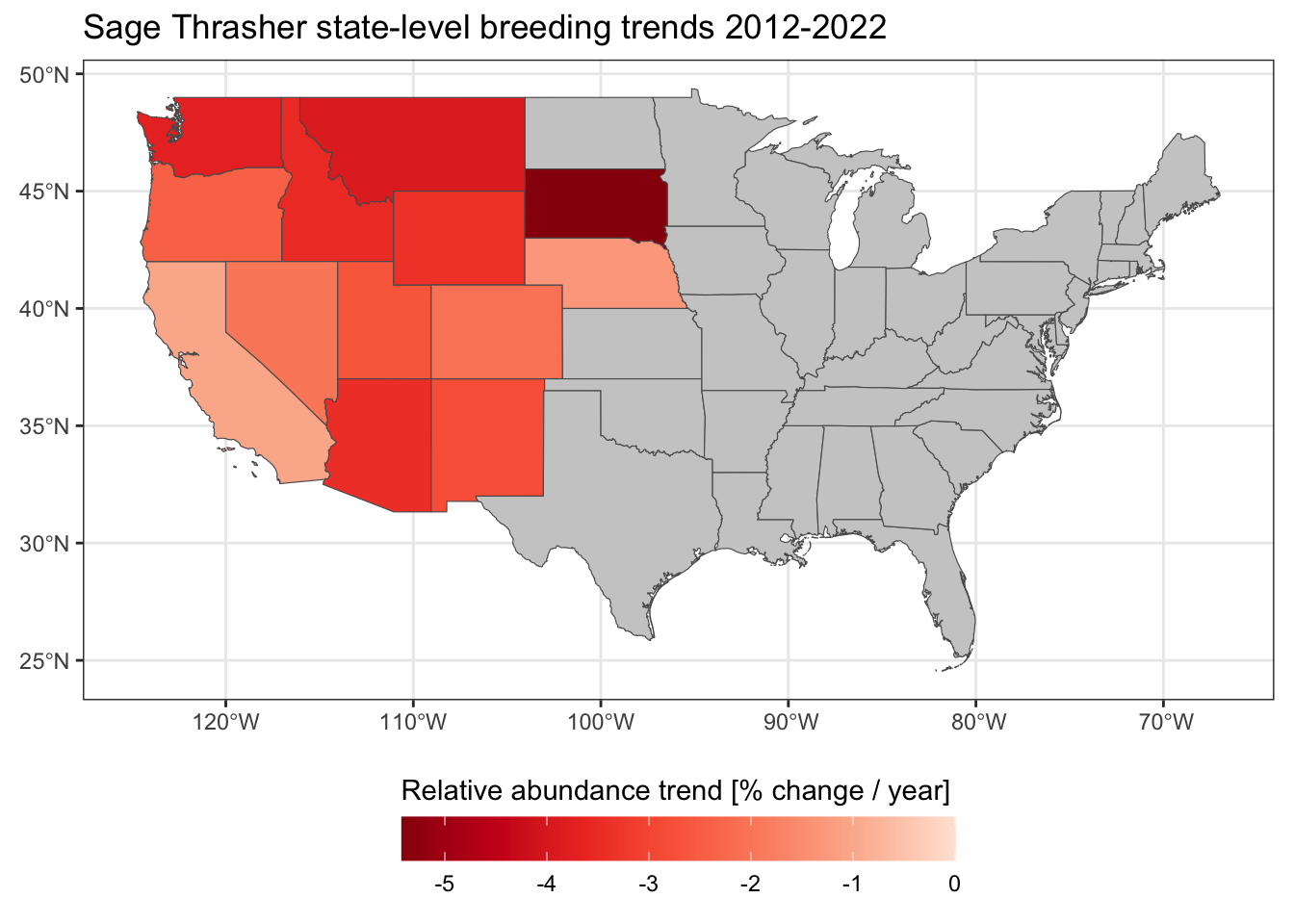
Based on these data, Sage Thrasher populations appear to be in decline throughout their entire range; however, some states (e.g. South Dakota) are experiencing much steeper declines than others (e.g. California).
2.6.2 Multi-species trends
In some cases, we may be interested in the trend for an entire community of birds, which can be estimated by calculating the cell-wise mean trend across a suite of species. For example, the eBird Trends Data Products contain trend estimates for three species that breed in sagebrush: Brewer’s Sparrow, Sagebrush Sparrow, and Sage Thrasher. We can calculate an average trend for this group of species, which will provide an estimate of the trend in the sagebrush bird community. First let’s look at the model information to ensure all species are modeled for the same region, season, and range of years.
sagebrush_species <- c("Brewer's Sparrow", "Sagebrush Sparrow", "Sage Thrasher")
trends_runs %>%
filter(common_name %in% sagebrush_species)
#> # A tibble: 3 × 10
#> species_code common_name trends_season trends_region trends_start_year
#> <chr> <chr> <chr> <chr> <dbl>
#> 1 brespa Brewer's Sparrow breeding north_america 2012
#> 2 sagspa1 Sagebrush Sparrow breeding north_america 2012
#> 3 sagthr Sage Thrasher breeding north_america 2012
#> # ℹ 5 more variables: trends_end_year <dbl>, trends_start_date <chr>,
#> # trends_end_date <chr>, rsquared <dbl>, beta0 <dbl>Everything looks good, so we can proceed to compare trends for these species. Next we need to download the trends data for these species. Note that since we’ve already downloaded the Sage Thrasher data above it won’t be re-downloaded here.
ebirdst_download_trends(sagebrush_species)Now we can load the trends and calculate the cell-wise mean to determine the average trend for the three species.
trends_sagebrush_species <- load_trends(sagebrush_species)
# calculate mean trend for each cell
trends_sagebrush <- trends_sagebrush_species %>%
group_by(srd_id, latitude, longitude) %>%
summarize(n_species = n(),
abd_ppy = mean(abd_ppy, na.rm = TRUE),
.groups = "drop")
head(trends_sagebrush)
#> # A tibble: 6 × 5
#> srd_id latitude longitude n_species abd_ppy
#> <int> <dbl> <dbl> <int> <dbl>
#> 1 234764 52.5 -118. 1 -8.61
#> 2 234765 52.5 -117. 1 -7.91
#> 3 234766 52.5 -117. 1 -6.30
#> 4 236265 52.2 -118. 1 -0.521
#> 5 236266 52.2 -117. 1 -6.90
#> 6 236267 52.2 -117. 1 -6.56Finally, let’s make a map of these sagebrush trends, focusing only on those cells where all three species occur.
# convert the points to sf format
all_species <- trends_sagebrush %>%
filter(n_species == length(sagebrush_species)) %>%
st_as_sf(coords = c("longitude", "latitude"),
crs = 4326)
# make a map
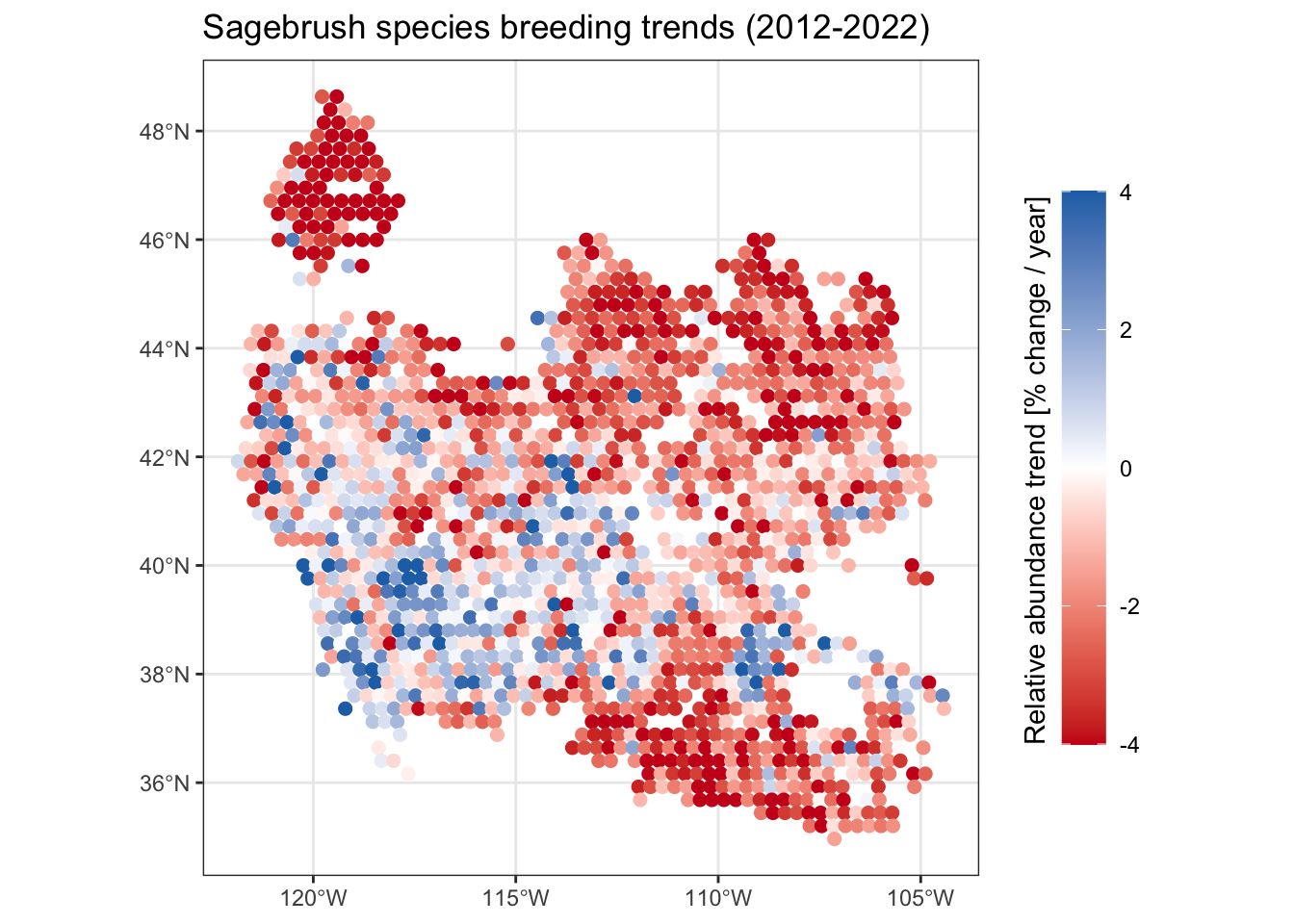
ggplot(all_species) +
geom_sf(aes(color = abd_ppy), size = 2) +
scale_color_gradient2(low = "#CB181D", high = "#2171B5",
limits = c(-4, 4),
oob = scales::oob_squish) +
guides(color = guide_colorbar(title.position = "left", barheight = 15)) +
labs(title = "Sagebrush species breeding trends (2012-2022)",
color = "Relative abundance trend [% change / year]") +
theme_bw() +
theme(legend.title = element_text(angle = 90))